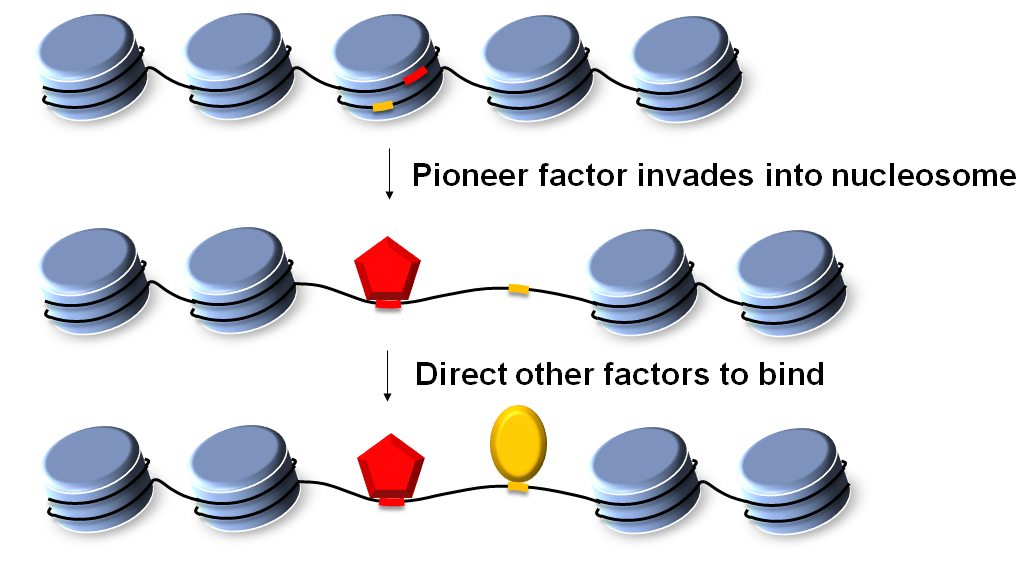
Chromatin is densely packed with nucleosomes, which limits the accessibility for most transcription factors. A special group of transcription factors can invade into compact chromatin and generate nucleosome-depleted regions (NDRs) near their binding sites. These factors, commonly referred to as “pioneer factors”, play essential roles in gene regulation by directing the binding of other transcription factors and allowing them to activate transcription. During development, pioneer factors function as “master regulators” to direct cell fate. Mis-regulation in pioneer factors are linked to cancer and developmental diseases. These findings raise the exciting possibility of using pioneer factors to generate cells with specific fates for regenerative therapy or as cancer drug targets. However, only a handful of pioneer factors are known in selected cell types; the properties that distinguish pioneer factors with canonical transcription factors are not clear; the mechanism of nucleosome invasion is also not well understood. Here, we developed a novel high-throughput assay, MNase over Integrated Synthetic Oligoes (MNase-ISO), to systematically measure the nucleosome-displacing activities among 104 yeast transcription factors. We identified a small subset as pioneer factors. Comparison between pioneer and non-pioneer factors indicate that abundance and strong DNA binding plays a significant role in pioneering activity. Incorporation of these pioneer factors into a thermodynamic model allows us to predict nucleosome positioning over the native yeast genome with high precision. This model can correctly predict ~65% of genome-wide NDRs.
Related publication:
- Thermodynamic Modeling of Genome-wide Nucleosome Depleted Regions in Yeast
Kharerin H & Bai L (2021)
Plos Computational Biology, 17(1): e1008560.
- Systematic Study of Nucleosome-Displacing Factors in Budding Yeast.
Yan C, Chen HY, & Bai L (2018)
Molecular Cell, 71(2): 294–30.

