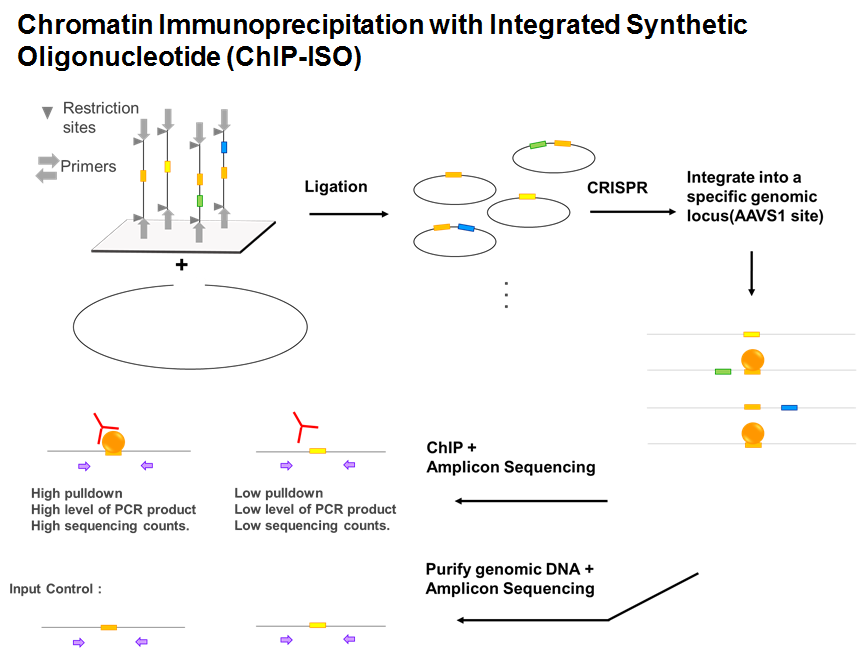
In higher eukaryotes, transcription factors only bind to a small fraction of their binding motifs across the genome, partially because of the inhibition of nucleosomes. Pioneer factors, however, are insensitive to nucleosome barrier and can bind to nucleosomal DNA. However, the fraction of motifs occupied by pioneer factors in the genome is still far from 100%. A well-characterized pioneer factor, FoxA1, for example, only binds to 10-20% of their strong consensus motifs in the genome. Many chromatin and sequence features can potentially contribute to this phenomenon, including DNA shape, co-factors, histone modifications, nuclear localization, etc. Here, we designed a high-throughput assay named Chromatin Immunoprecipitation with Integrated Synthetic Oligonucleotides (ChIP-ISO) to systematically dissect local sequence features affecting the binding of a classic PF, FoxA1, in A549 human lung carcinoma cells. This method involves integrating thousands of synthetic sequences containing FoxA1 motifs into a fixed genomic locus, followed by FoxA1 ChIP and next-generation amplicon sequencing. We found that within the same sequence background, FoxA1 binding is strongly affected by its motif strength, motif orientation, number of motifs and co-binding TFs including Fos/Jun and CEBPB, among which Fos/Jun is particularly important for enhancing FoxA1 binding. Finally, by moving sequences originated from genomic loci with different epigenetic marks to the same chromatin background and measuring FoxA1 binding, we showed that FoxA1’s binding specificity is more determined by the local sequence than the chromatin background. In summary, our study provides insights of the genetic rules underlying pioneer factor binding specificity and reveals a potential mechanism to regulate its binding during cell differentiation.

